Difference between revisions of "Multi-Genome Tutorial"
From GenPlay, Einstein Genome Analyzer
(→Files) |
(→Files) |
||
| Line 149: | Line 149: | ||
*[http://www.genplay.net/library/Human/Multi-Genome/hg18tohg19_tutorial_sv.vcf.gz.tbi Indexed VCF file (Tabix)] | *[http://www.genplay.net/library/Human/Multi-Genome/hg18tohg19_tutorial_sv.vcf.gz.tbi Indexed VCF file (Tabix)] | ||
*[http://www.genplay.net/library/Human/hg19/Gene_Annotation/RefSeq_From_UCSC_04-23-10(hg19).bed Refseq BED file for GRCh37/hg19] | *[http://www.genplay.net/library/Human/hg19/Gene_Annotation/RefSeq_From_UCSC_04-23-10(hg19).bed Refseq BED file for GRCh37/hg19] | ||
| − | *[http://www.genplay.net/library/Human/hg18/Gene_Annotation | + | *[http://www.genplay.net/library/Human/hg18/Gene_Annotation/RefSeq_From_UCSC_04-23-10(hg18).bed Refseq BED file for NCBI36/hg18] |
=== Steps === | === Steps === | ||
Revision as of 15:57, 13 September 2011
Contents
Getting started
Introduction
To create a multi-genome session, users must first load VCF files. VCF files are files that describe all the differences between a reference genome and a particular genome.
The first part of this tutorial presents how to set up a multi-genome project, especially how to load VCF files. The second part concerns loading data tracks. Data tracks must be mapped to one of the loaded genomes. Finally, we will describe how to highlight information from VCF files.
The Welcome screen
The welcome screen is the first screen of GenPlay-MG and allow user to create or to load a project.
New Project
In order to create a new project, users must give it a name as shown in Figure 1.
The second step is to choose a reference genome. Users can choose it using the different list according to the clade, the genome and the assembly (Figure 2).
Several chromosomes are available for each assembly but users can choose to select only some of them.
To open the chromosome chooser (Figure 3), users have to click on the button next to the assembly name.
The third and last step is to choose between a Simple Genome Project and a Multi Genome Project. This tutorial is about multi genome project, after having checked this option, the welcome screen should be as the one shown in Figure 4.
Load Project
Coming soon
(Option unavailable in the beta version)
VCF Files
Description
VCF files describe differences between two genomes. Usually, it concerns differences between a genome of interest and the reference genome used for the mapping process. There are three different types of VCF files. They differ by the type of variants they describe:
- Indels
- SNPs
- SV (Structural Variation)
A complete description of VCF files is given on the 1000 genomes project website:
VCF (Variant Call Format)
Encoding Structural Variants in VCF
Tabix
Introduction
VCF files contain a lot of information which makes the scanning (loading) processes longer.
In order to increase the scanning efficiency, VCF files are indexed with Tabix.
Tabix manual reference pages
Tabix download
VCF files indexation method
Each VCF files must be first compress to a BGZF (.bgz file) format. Tabix provides a tool to perform the compression.
After compression, VCF files must be indexed using the associated command.
Once Tabix is installed, two commands are necessary to perform the indexation.
Available commands from the Tabix folder:
bgzip -f VCF_PATH;
tabix –p vcf VCF_PATH;
For example, a VCF file named my_vcf.vcf located in the same folder as Tabix can be indexed with the following commands (Figure 5):
bgzip -f ./my_vcf.vcf;
tabix –p vcf ./my_vcf.vcf.gz;
Note: the first command replaces the current VCF file by the compressed VCF file (.vcf.gz). The second command creates the indexed VCF file in the current folder (.vcf.gz.tbi).
More options are available on Tabix manual reference pages.
VCF files loading
The VCF Loader
Introduction
The VCF Loader is the most important part of multi-genome project settings. It allows users to load all necessary VCF files and to define how to extract information from them. It appears when users click on the "Edit" button from the welcome screen.
The Figure 6 shows an empty VCF Loader screen.
To understand the loading process, it is necessary to know that VCF files can contain information pertaining to one type of variant (SNP, indel or SV) for multiple individuals and that the genome of one individual is often described by 3 VCF files respectively describing the SNPs,the indels and the SV.
To load the complete genome of one or several individuals GenPLay-MG must therefore open several VCF files and organizes them according to genomes and group of genomes: In this example, a group of genome will represent a family, and a genome a member of the family. Each row in the table represents a genome and a VCF file (and therefore a type of variant).
If a project (a session) involves 2 families with 3 individuals each and if all 3 different types of VCF file (indels, SNPs, SV) are available for each individual, 18 lines will have be filled (2 families x 3 individuals x 3 VCF types). If user wants to compare individuals from the same family, he still has to fill group cells (with the same name).
Note: GenPlay-MG does not use directly the VCF file, it uses a compress version of it (.gz). Moreover, GenPlay-MG also needs the compress VCF file to be indexed with Tabix. Both file versions must be in the same folder and must have the same name, only file extensions differ (.gz and .tbi).
The user can add or remove rows.
When user has done, the "ok" button will valid settings but the "cancel" button and closing the window will cancel every modification.
Columns description
Group
Users can gather genomes by group. Group names are used to distinguish genomes and to perform some specific functionalities.
Genome
The Genome column allows users to associate an alias to the selected genome. This alias will appear in GenPlay-MG and can be useful because genome names in VCF files are often non descriptive numbers that can be hard to remember.
Type
The Type column contains the different types of VCF files. At the current time GenPlay-MG recognizes three types of VCF files:
- Indels
- SNPs
- SV (Structural Variations)
File
This column refers to the VCF file path. Once loaded, the raw name column is automatically filled with every raw genome name contained in the selected VCF file.
Raw name(s)
The Raw name(s) column list is automatically filled when a VCF file has been chosen. That list contains every genotype headers contained inside the selected VCF file. Because Genome names might be difficult to remembers, GenPlay-MG offers users the option of adding another name (an alias) using the Genome column.
Columns edition
Group, Genome and File column have their own editable list. To edit a column, user has to go to Column list edition on the bottom left of the VCF Loader (Figure 7).
Once the user has clicked on the Edit button, a new window appears (as shown on Figure 8 for the group column) with a plus and a minus button in order to add or remove elements from the list. Plus button will show a new dialog allowing users to write a short text.
In the case of the File column, the plus button opens a file chooser.
That way, users can set up all columns before starting (or at the same time) to fill the table.
Note: The Type column contains static value, it is not editable. The Raw name(s) column is automatically filled with genome name from the selected VCF file, that column cannot be edited manually.
Import/Export
Once a project has been set up, it can be saved using the import/export function. Pressing the export button saves an XML files to the hard drive. This XML file can then be imported to reload the project.
The XML file structure is simple. Each row are stored in row mark containing every attribute names such as group, genome, type, file and raw_name. The settings file is formatted as shown in Figure 9.
Note: If the user moves the VCF files or changes one of its genotype headers, the XML file will not work anymore. User has to modify file and/or raw_name attribute values.
Tracks loading
Once a multi-genome project has been created, GenPlay creates a meta-genome that is the sum of all the loaded genomes and is capable of converting the coordinates of any data files into the meta-genome coordinates. GenPLay can therefore load data files (tracks) mapped in the coordinates of any of the loaded genomes. Of course, when loading a file, user must specify which genome was used for the mapping.
When loading a track, GenPlay displays a list showing every loaded genome below the window allowing user to load the files. Once the information is entered, GenPlay transforms the coordinates in the file into the coordinates of the meta-genome using the differences information of the specified genome.
Displaying information
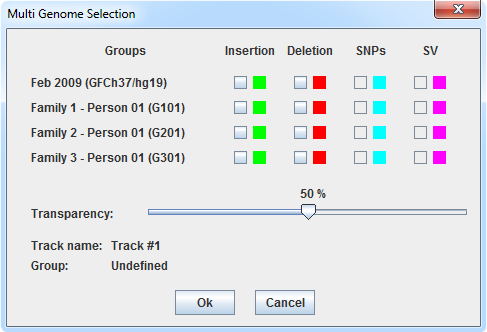
In order to display variants information related to genome differences, each track has its own window settings. When user right-clicks on the track handler (on the left of the track) of an empty tracks, and clicks on Multi Genome Stripes the window below appears.
In this example,, there are three families with one member each. Genomes have been mapped on the reference genome GRCh37/hg19. It is possible to show on the selected track information such as insertion, deletion, SNPs and structural variants. User can define the colors (by clicking on the colored squared) and also define stripes transparency using the slider.
If a data file concerning Person 01 of the first family has been loaded, the window looks like the one on Figure 11.
Firstly, all stripes related to variants of the selected genome appear (In this case the insertion and the deletion for person 1, family 1). Secondly, all insertions in the genome of family 1 are shown as black stripes on all other genomes (black stripes are synchronization marker that have been introduced in the meta-genome to be able to display multiple genome at the same time. The black stripes represent insertions in genome others than in the current genome. User can modify stripes visualization using this panel.
Conversion between NCBI36/hg18 and GRCh37/hg19
Description
This tutorial will explain how to display at the same time tracks mapped on genome assembly NCBI36/hg18 or GRCh37/hg19. In the example, user will be able to see all the modifications on the NCBI36/hg18 genome leading to the GRCh37/hg19 reference genome.
Files
- XML settings file
- VCF file
- Indexed VCF file (Tabix)
- Refseq BED file for GRCh37/hg19
- Refseq BED file for NCBI36/hg18
Steps
Project settings
Project name
User must choose a name for a new project; here the name is GenPlay-MG – Reference genome tutorial (Figure 1).
Project assembly
According to bed files provided in this tutorial, the reference genome is GRCh37/hg19. User has to select the mammal clade, the human genome and the Feb 2009 (GRCh37/hg19) assembly as in Figure 2.
Chromosome selection
The VCF file is about Structural Variants and contains information for chromosomes 1 to 22 and chromosomes X and Y. User can select to load on or more chromosomes by clicking on the settings button next to the assembly name (Figure 3).
VCF Loading
Manually
To load VCF files , users must first fill the column lists and then select from the list the appropriate data. The VCF Loader appears after clicking on the Edit button from the welcome screen. The bottom left part of the VCF Loader contains the Column list edition section. User has to select a column and click on Edit button in order to show the associated list.
Only one VCF file is going to be loaded for this tutorial. The VCF file contains differences between the reference genome NCBI36/hg18 and the reference genome GRCh37/hg19.
Group column
This tutorial compares reference genome; a generic group name can be Reference genome.
On the Group name list editor, user clicks on the plus button to show the input text box and fills it (Figure 4).
The Group name list editor should looks like the Figure 5 below.
Once the values has been added to the list, it can be saved by closing the "Group name list editor window"
value: Reference genome
Genome column
The genome name is an Alias for the selected raw name. In this tutorial, the genome name is going to be Hg18.
On the Genome name list editor, user clicks on the plus button to invoke the input text box and fills it (Figure 6).
The Genome name list editor should looks like the Figure 7 below.
Once the values has been added to the list, it can be saved by closing the "Genome name list editor window"
value: Hg18
Type column
This field cannot be edited by the users. The provided VCF file is a Structural Variant type, user therefore has to choose SV (Figure 8).
value: SV
File column
Once the VCF file is downloaded, user has to open the File list editor, user clicks on the plus button to show the file chooser dialog and choose the VCF file according to its location.
value: VCF path
Raw name(s) column
The raw name list is automatically filled. In the case of this tutorial there is only one genome: NCBI36 (Figure 10).
value: NCBI36
Again, value is saved by closing the windows
Import XML settings
In order to set the project with ease, user can import the settings using the XML file above. Please be careful about the VCF path, user must changes it directly on the xml file if he wants to use the import function.
Conclusion
Finally, the screen should be like the one on Figure 11.
Conclusion
The welcome screen should finally be similar to the Figure 12.
The "Create" button will create the project and will run the synchronization.
GRCh37/hg19 genes loading
To load a file, user has to do a right click on the left part of the track. Then to choose "Load Gene Track", a file chooser appears to select the file given in this tutorial. After having chosen the BED file, a new selection box appears (Figure 13).
This box asks which genome is related to the BED file. Here, user has to choose "Feb 2009 (GFCh37/hg19)" option because the BED file contains information about that genome. Gene file for GRCh37/hg19 reference has been loaded.
NCBI36/hg18 genes loading
The same operation as loading a gene files for GRCh37/hg19 reference genome. The only step changing is to choose the "Reference genome - hg18 (NCBI36)" option after the BED file selection (Figure 14)
Conclusion
User can navigate into the different chromosomes and visualizes differences between both genomes using the stripes. All genes are perfectly synchronized and are display according to the meta-genome coordinates.
The Figure 15 shows an example of the result of this tutorial. It is possible to see deletions (in red) and insertions (in green) in the NCBI36/Hg18 reference genome compare to the GCh37/Hg19 reference genome.
Chromosome: chr1
Position: 143,822,670